ScanNeo2: a comprehensive workflow for neoantigen detection and immunogenicity prediction from diverse genomic and transcriptomic alterations
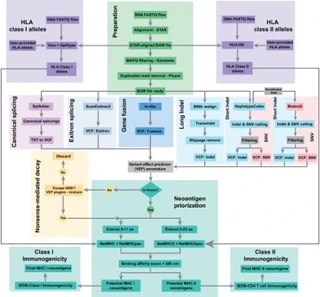
Neoantigens, tumor-specific protein fragments, are invaluable in cancer immunotherapy due to their ability to serve as targets for the immune system. Computational prediction of these neoantigens from sequencing data often requires multiple algorithms and sophisticated workflows, which are currently restricted to specific types of variants, such as single-nucleotide variants or insertions/deletions. Nevertheless, other sources of neoantigens are often overlooked.
We introduce ScanNeo2, an improved and fully automated bioinformatics pipeline designed for high-throughput neoantigen prediction from raw sequencing data. Unlike its predecessor, ScanNeo2 integrates multiple sources of somatic variants, including canonical- and exitron-splicing, gene fusion events, and various somatic variants. Benchmarking demonstrates that ScanNeo2 accurately identifies neoantigens, providing a comprehensive and more efficient solution for neoantigen prediction.
