OctopuSV and TentacleSV: a one-stop toolkit for multi-sample, cross-platform structural variant comparison and analysis
Motivation
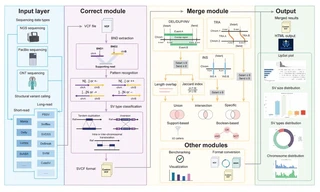
Structural variants (SVs) influence gene regulation, disease progression, and diagnostics, yet integrating SV calls across platforms remains difficult due to inconsistent annotations, limited merging flexibility, and fragmented workflows. Ambiguous breakend (BND) annotations, which comprise many variant calls, are often discarded or misclassified, hindering variant characterization. Existing tools lack advanced merging operations essential for precise identification of disease-specific or somatic variants across samples or patient groups. Additionally, current SV analysis pipelines require extensive manual intervention and complex parameter tuning, compromising reproducibility and scalability. Addressing these gaps is crucial for improving the accuracy, interpretability, and clinical utility of SV analyses.
Results
We developed OctopuSV and TentacleSV to address these long-standing challenges in SV analysis. OctopuSV features a specialized BND correction module that converts ambiguous BND annotations into canonical SV types, recovering important variants that are often overlooked by existing tools. Additionally, it provides advanced set operations (difference, complement, custom-defined) that enable sophisticated variant filtering without programming expertise, critical for identifying tumor-specific SVs or variants unique to specific sample groups. TentacleSV completes our solution by automating the entire SV analysis process from raw sequencing data to high-confidence callsets, ensuring consistency and reproducibility across projects. Benchmarking across short-read and long-read platforms showed superior F1 score, complete SV type consistency compared to existing tools. Our framework enables experimental biologists and clinical researchers to perform sophisticated analyses ranging from cancer subtype-specific SV identification to multi-sample comparative studies without requiring specialized programming skills.
Availability and implementation
All codes are available at github.com/ylab-hi/OctopuSV and github.com/ylab-hi/TentacleSV.
DOI: https://doi.org/10.1093/bioinformatics/btaf599
Journal page: https://academic.oup.com/bioinformatics/article/41/11/btaf599/8307122
Code: https://github.com/ylab-hi/OctopuSV
Pipeline: https://github.com/ylab-hi/TentacleSV
