I received my Ph.D. in Bioinformatics at Huazhong Agricultural University in Jun 2021. With the generous guidance of Prof. Jerri Bartholomew, that experience sparked my lasting interest in host-parasite interactions and evolutionary genomics. My research background is in multi-omic insights (genomics, transcriptomics, and proteomics) into the early animal genome evolution and genetic basis of their phenotypic adaptation. And I am very interested in developing multimodal pipelines to discover fundamental principles that control phenotype.
During my PhD, I investigated the evolutionary genomics of the myxozoans, micro-meter sized parasitic cnidarians, and found a new model of parasite evolution – mosaic evolution (BMC Biology, 2022). I developed a customized comprehensive proteomic reference database (CCPRD) pipeline, which has greatly improved the efficiency and accuracy of proteomic research in non-model organisms. I also applied proteomics, algorithm development, and quantitative genetic analysis to demonstrate that nematocysts may be a key determinant of the adaptive success of cnidarians. I also modelled the relationship between the evolutionary mode of cnidarians and palaeo-environmental change and found that the diversification of cnidarians is predominantly uncoupled from palaeoclimate.
I have authored and co-authored 25 peer-reviewed publications, including works published on Science Advances, BMC Biology, Biology, Journal of Experimental Biology, and Parasites & Vectors (seven papers are first author and three more in preparation).
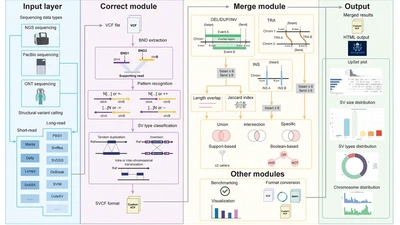
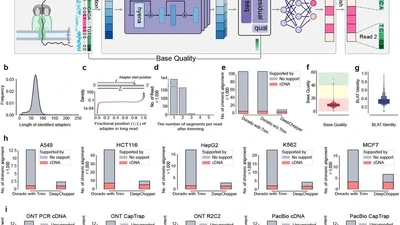
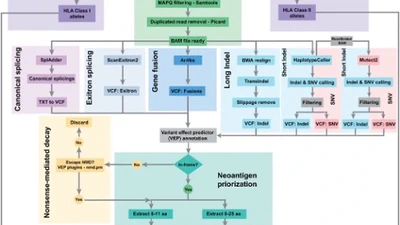
My long-term goal is to become an independent academic researcher advancing the use of multi-omics and AI-driven methods in biomedical genomics. While my Ph.D. work focused on parasitic cnidarians, my current research as a postdoctoral fellow in Dr. Rendong Yang’s lab at the Feinberg School of Medicine, Northwestern University, centers on the structural variations of cancer genomes and single-cell omics. I develop long-read and deep learning based tools to decode the regulatory complexity of cancer genomes, with the aim of enabling earlier detection, more precise target discovery, and personalized therapies - hoping that these approaches will help bridge the gap between genomic insights and practical tools for cancer diagnosis and treatment.